Table of Contents
The Network Comparison Toolkit, or NCT was designed to be an extensible platform for comparing biological networks. The NCT is written in pure Java 1.5 and it now represents the canonical versions of the various algorithms used for comparisons. The NCT has its roots in the PathBLAST project and implementations of the underlying algorithims in C, Python, and Java. The motivation for the reimplementation of the algorithms is the development of software that is modular, easily maintained and easily extended for future research.
NCT is heavily modularized meaning it is decomposed into separate Java classes that perform specific duties. The classes interact with one another through clearly defined interfaces that allow new algorithms and scoring methods to be implemented without rewriting unrelated code. The intended use of the NCT classes is exemplified in the NetworkBlast program. The fundamental approach is:
- Construct the input graph objects.
- Create the scoring model objects.
- Apply search algorithms to the graphs using the scoring models.
- Capture the output.
There are several classes defined for each step described which allows programmers to mix and match classes to easily meet their specific needs.
NCT is broken down into several packages, but there are four primary types of object that we care about: Graphs, Search Algorithms, Scoring Models, and Utilities. In addition, specific applications have also been implemented using the NCT modules. These modules are described in the following chapter.
Table of Contents
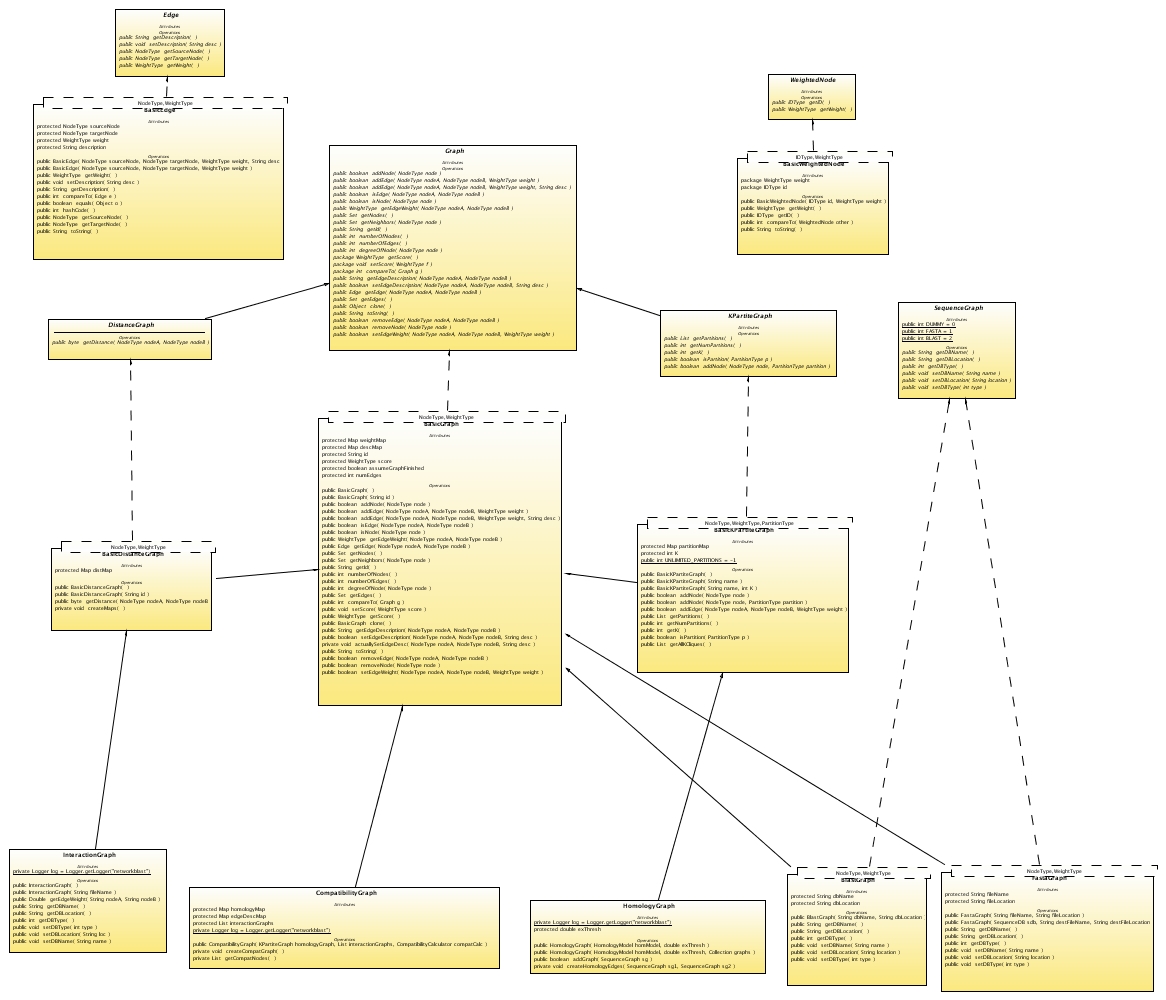
The common currency of NCT is the Graph interface found in the nct.graph package. The interface defines how programmers should interact with graph objects. The Graph interface has several extensions that expand on the capabilities of a basic graph such as the KPartiteGraph which defines the methods used for interacting with k-partite graphs. Basic implementations of all the Graph interfaces have been provided in the nct.graph package.
The standard graph interfaces and definitions are general and do not contain biological semantics. This means that the standard graph classes and interfaces could be used as a general graph library.
Graph/BasicGraph
The graph class defines all of the basic graph operations such as addNode, addEdge, getNeighbors, numberOfNodes, numberOfEdges, and the like. A basic implementation of the Graph interface exists and is called BasicGraph.
DistanceGraph/BasicDistanceGraph
The DistanceGraph interface extends the Graph interface by adding a getDistance method that is intended to calculate the shortest path between two nodes. The BasicDistanceGraph extends BasicGraph and adds a getDistance method which assigns a distance of 1 to neighbors, a distance of 2 to neighbors of neighbors and 3 to all other nodes.
KPartiteGraph/BasicKPartiteGraph
The KPartiteGraph interface defines several methods for creating and defining k-partite graphs. The BasicKPartiteGraph implementation extends the BasicGraph class and overrides methods that don't respect partitisions such as addNode(node), which is replaced with addNode(node,partition).
SequenceGraph/FastaGraph/BlastGraph
SequenceGraph is an interface that associates biological sequences with the nodes of the graph. The Fasta and Blast implementations simply represent two types of sequence database that can be used.
The network comparision graphs are extensions of the standard graphs that are designed for specific network comparison tasks.
CompatibilityGraph
HomologyGraph
InteractionGraph
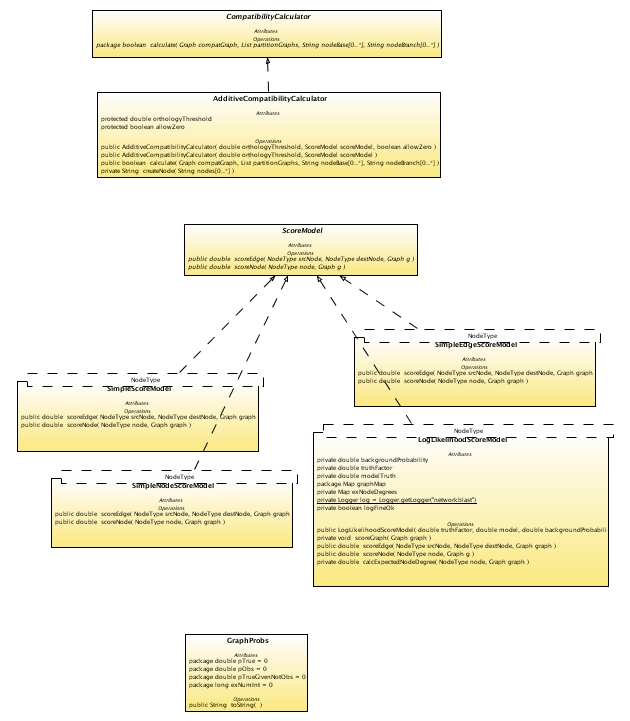
This module contains various interfaces that define mechanisms for calculating various graph scores. The scoring algorithm classes are contained in the nct.score package.
The ScoreModel interface defines two methods for returning the score of a node or edge on a graph. How those scores are calculated is left to the implementation of the interface (class).
LogLikelihoodScoringModel
SimpleScoringModel
SimpleEdgeScoringModel
SimpleNodeScoringModel
There are several alternative techniques for determining whether nodes in interaction graphs should be aligned. The CompatibilityCalculator provides an interface that lets different compatibility determination schemes to be explored.
AddititiveCompatibilityCalculator
....
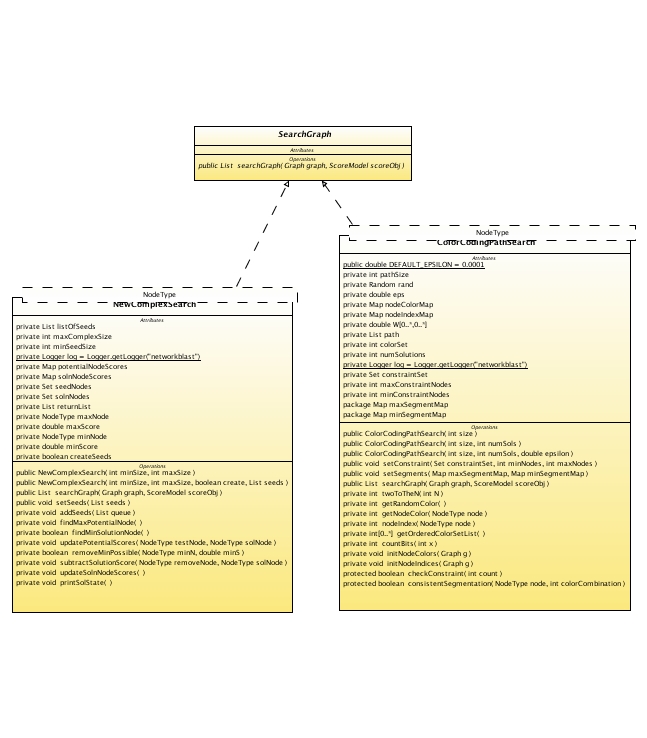
The search algorithm module is nct.networkblast.search.
ColorCodingPathSearch
GreedyComplexSearch
The services package is comprised of several sub-packages. The intent of the package is isolate activities that are not core graph comparison duties in a separate package. Examples include the calculation of homology e-values between sequences, the creation of interaction graphs, the creation of synonym databases, access to sequence databases, and the like. In each case an interface is provided that defines how new software should interact with external services.
synonyms
homology
interactions
sequences
filters
The filter package provides a simple interface for filtering a collection of Graphs. Examples of the filters provided are the SortFilter and the DuplicateThresholdFilter. The SortFilter merely sorts a collection of Graphs based on the Graph's implementation of the Comparable interface. The DuplicateThresholdFilter filters out Graphs that have a certain percentage (the threshold) of nodes and edges in common.
output
Classes for writing output in useful ways (e.g. graphs into the SIF format).
cytoscape
xml
parsers
NetworkBlast, as the name suggests, is where the classes specific to the NetworkBlast/PathBlast algorithms reside. This package contains the CompatibilityGraph that represents the network alignment and the HomologyGraph used for calculating that alignment.
Central to the NetworkBlast package is the NetworkBlast class itself. This class contains the main method that is called upon invocation of the networkblast program. The main method contains the actual code that reads the input, constructs the input graphs, defines the search and scoring classes used for the alignment, and shows how the output is generated. If there is any confusion about how certain classes relate to one another, a look at the main method might help clarify things.
Whenever possible we strongly recommend that you code to the graph interfaces and not the specific implementations. For example:
// recommended usage Graph<String,Double> g = new BasicGraph<String,Double> // NOT recommended BasicGraph<String,Double> g = new BasicGraph<String,Double> // recommended method definition public Graph<String,Double> getGraph(); // NOT recommended public BasicGraph<String,Double> getGraph();
Coding to the interface provides flexibility to other users of your code to change underlying implementations without having to alter your code.