Perhaps the most important consequence of the Human Genome Project is that it is pushing scientists toward a new view of biology what we call the systems approach. Systems biology does not investigate individual genes or proteins one at a time, as has been the highly successful mode of biology for the past 30 years. Rather, it investigates the behavior and relationships of all of the elements in a particular biological system while it is functioning. These data can then be integrated, graphically displayed, and ultimately modeled computationally.
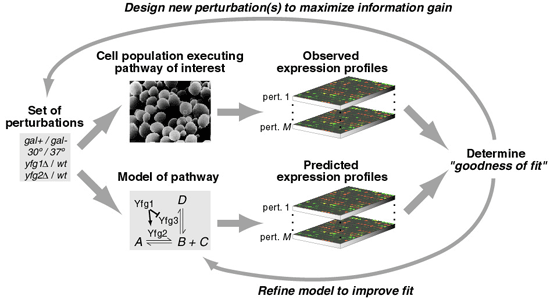
As illustrated above, Systems Biology involves several distinct steps:
(i) Define all of the genes in the genome and the subset of genes, proteins, and other small molecules constituting the pathway of interest. If possible, define an initial model of the molecular interactions governing pathway function, drawn from previous genetic and biochemical research.
(ii) Perturb each pathway component through a series of genetic (e.g. gene deletions or overexpressions) or environmental (e.g. changes in growth conditions or temperature) manipulations. Detect and quantify the corresponding global cellular response to each perturbation with technologies for large-scale mRNA- and protein-expression measurement.
(iii) Integrate the observed mRNA and protein responses with the current, pathway-specific model and with the global network of protein-protein, protein-DNA, and other known physical interactions.
(iv) Formulate new hypotheses to explain observations not predicted by the model. Design additional perturbation experiments to test these and iteratively repeat steps (ii), (iii), and (iv).
We are working in close collaboration with our colleagues at the Institute for Systems Biology to apply this strategy to the study of human and model organisms. Surprisingly, many of the obstacles to achieving this strategy are computational in nature, and they are the focus of ongoing research in the Ideker lab. |
